Research

IR Bactyping is an accurate (>95%), easy to implement and low-cost strain typing tool based on a biochemical fingerprint obtained by Fourier-Transform Infrared spectroscopy that can reduce by >75% the time-to-response (less than 24h) and by 90% the cost of analysis compared to reference typing methods. Because we have proprietary spectral databases, we are able to identify specific strain types that represent main and well-defined lineages involved in local or nationwide epidemics in multiple countries. Precise and timely strain type information is crucial to early identify outbreak events and particular high-risk clones and will contribute to support real-time surveillance, precise and effective infection control measures in healthcare settings.
Rodrigues C, Sousa C, Lopes JA, Novais Â, Peixe L. A Front Line on Klebsiella pneumoniae Capsular Polysaccharide Knowledge: Fourier Transform Infrared Spectroscopy as an Accurate and Fast Typing Tool. mSystems. 2020 Mar 24;5(2):e00386-19.
Novais Â, Freitas AR, Rodrigues C, Peixe L. Fourier transform infrared spectroscopy: unlocking fundamentals and prospects for bacterial strain typing. Eur J Clin Microbiol Infect Dis. 2019 Mar;38(3):427-448. doi: 10.1007/s10096-018-3431-3.
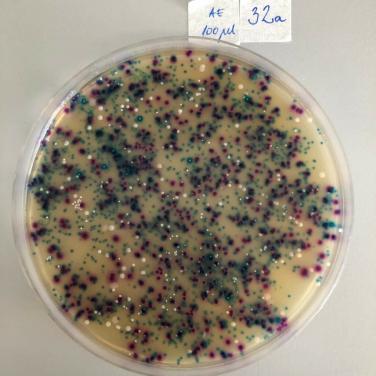
Members of CCP have been enrolled in the accurate identification, characterization, and conservation of strains from different bacterial genus (Bacillus, Lactobacillus, Limosilactobacillus, Citrobacter) and sources (health product, water, human faecal and urinary samples). Branquinho R, Sousa C, Osório H, Meirinhos-Soares L, Lopes J, Carriço JA, Busse HJ, Abdulmawjood A, Klein G, Kämpfer P, Pintado ME, Peixe LV. Bacillus invictae sp. nov., isolated from a health product. Int J Syst Evol Microbiol. 2014 Nov;64(Pt 11):3867-3876.
Rocha J, Botelho J, Ksiezarek M, Perovic SU, Machado M, Carriço JA, Pimentel LL, Salsinha S, Rodríguez-Alcalá LM, Pintado M, Ribeiro TG, Peixe L.
Lactobacillus mulieris sp. nov., a new species of Lactobacillus delbrueckii group. Int J Syst Evol Microbiol. 2020 Mar;70(3):1522-1527.
Ksiezarek M, Ribeiro TG, Rocha J, Grosso F, Perovic SU, Peixe L. Limosilactobacillus urinaemulieris sp. nov. and Limosilactobacillus portuensis sp. nov. isolated from urine of healthy women. Int J Syst Evol Microbiol. 2019 Jun;71(3).
Ribeiro TG, Gonçalves BR, da Silva MS, Novais Â, Machado E, Carriço JA, Peixe L. Citrobacter portucalensis sp. nov., isolated from an aquatic sample. Int J Syst Evol Microbiol. 2017 Sep;67(9):3513-3517.
Ribeiro TG, Clermont D, Branquinho R, Machado E, Peixe L, Brisse S. Citrobacter europaeus sp. nov., isolated from water and human faecal samples. Int J Syst Evol Microbiol. 2017 Jan;67(1):170-173.
Ribeiro TG, Izdebski R, Urbanowicz P, Carmeli Y, Gniadkowski M, Peixe L. Citrobacter telavivum sp. nov. with chromosomal mcr-9 from hospitalized patients. Eur J Clin Microbiol Infect Dis. 2021 Jan;40(1):123-131.
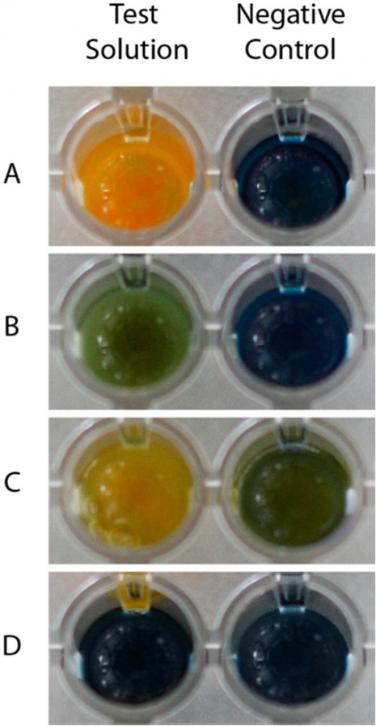
Quick, simple, and reliable methods are needed for laboratory detection of carbapenemases that are widely disseminated among Gram-negative bacteria, in order to improve the detection and surveillance of these clinically relevant bacteria in an epidemiological context. The Blue-Carba was proposed and validated for the detection of carbapenemase activity directly from bacterial cultures of Enterobacterales, Pseudomonas and Acinetobacter species.
Pires J, Novais A, Peixe L. Blue-carba, an easy biochemical test for detection of diverse carbapenemase producers directly from bacterial cultures. J Clin Microbiol. 2013 Dec;51(12):4281-3.
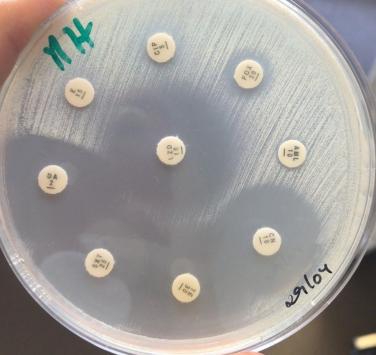
The emergence of antibiotic resistance creates a new challenge for public health, and there is no simple solution. CPP is hosting a unique collection of clinical isolates to study history and evolution of resistance in ESKAPE pathogens and many others. Resistance to antibiotics can be either intrinsic to a given bacterial species or acquired by genetic mutations and foreign DNA not present in the natural populations. The means to identify and genes coding for resistance to antibiotics are integrated in our pipelines of genome data analysis from CIP deposited strains. We are developing sophisticated methods to implement such a resistance gene prediction workflow in order to detect what is not already known, searching all possible acquired resistance genes and novel resistance signatures.
Freitas AR, Pereira AP, Novais C, Peixe L. Multidrug-resistant high-risk Enterococcus faecium clones: can we really define them? Int J Antimicrob Agents. 2021 Jan;57(1):106227.
Ribeiro TG, Novais Â, Branquinho R, Machado E, Peixe L. Phylogeny and Comparative Genomics Unveil Independent Diversification Trajectories of qnrB and Genetic Platforms within Particular Citrobacter Species. Antimicrob Agents Chemother. 2015 Oct;59(10):5951-8.
Gonçalves Ribeiro T, Novais Â, Machado E, Peixe L. Acquired AmpC β-Lactamases among Enterobacteriaceae from Healthy Humans and Animals, Food, Aquatic and Trout Aquaculture Environments in Portugal. Pathogens. 2020 Apr 9;9(4):273.
